 |
| Posters for the people! |
Download a copy of the poster here.
Until next time...

Bentley Historical Library Curation Team
The Bentley Historical Library's Mellon-funded ArchivesSpace-Archivematica-DSpace Workflow Integration project (2014-2016) united three Open Source platforms for more efficient creation and reuse of metadata and to streamline the ingest of digital archives. We continue to explore innovative archival practice and emerging technologies to curate our collections—read all about it here!
 |
| Posters for the people! |
The Digital Object record is optimized for recording metadata for digitized facsimiles or born-digital resources. The Digital Object record can either be single- or multilevel, that is, it can have sub-components just like a Resource record. Moreover, the record can represent the structural relationship between the metadata and associated digital files--whether as simple relationships (e.g., a metadata record associated with a scanned image, and its derivatives) or complex relationships (e.g., a metadata record for a multi-paged item; and additionally, a metadata record for each scanned page, and its derivatives). One or more file versions can be referenced from the Digital Object metadata record. The Digital Object record can be created from within a Resource record, or created independently and then either linked or not to a Resource record.
 |
| Unlike the actual episode from Season 3 of the A-Team, our big squeeze does not involve loan sharks, mob extortion bosses, breaking fingers, sniper assaults (phew!), or Irish pubs (or does it...). |
 |
| The plan. Ignore that stuff on the bottom left. |
 |
| We'll be making fairly extensive use of the user-defined portion of ArchivesSpace accession records. Has anyone changed the labels for these fields? If so, let us know! We're keen to explore this. |
| Another famous |
| WinDirStat |
| Bulk Extractor Viewer |
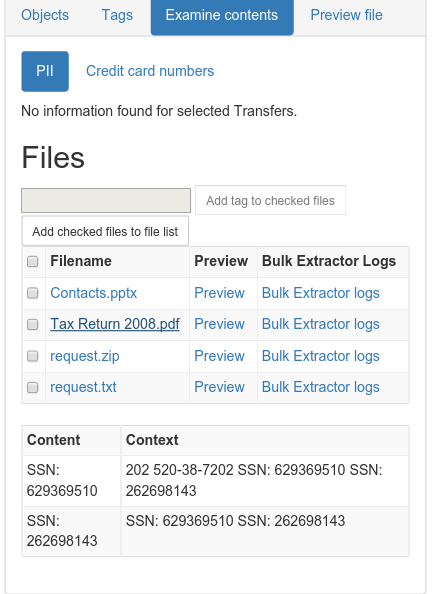 |
| SSNs identified! |
| Lexical Dispersion Plot for Words in U.S. Presidential Inaugural Addresses: This can be used to investigate changes in language use over time. |
 |
| This plot shows lexical diversity over time for the Michigan Technic, a collection we've digitized here at the Bentley, using Seaborn, a Python visualization library based on matplotlib. Not much of a trend either way, although I'm not sure what happened in 1980. |
| Not exactly an archival example, but one of my favorites nonetheless. The Largest Vocabulary in Hip Hop: Rappers, Ranked by the Number of Unique Words in their Lyrics. The Wu Tang Association is not to be trifled with. |
| Cumulative Frequency Plot for 50 Most Frequently Words in Moby Dick: these account for nearly half of the tokens. |
| Stanford Named Entity Recognizer |
| ePADD, a software package developed by Stanford University's Special Collections & University Archives, supports archival processes around the appraisal, ingest, processing, discovery, and delivery of email archives. This image previews it's natural language processing functionality. |
| Those lists of places could be geo-coded and turned into something like this, a map of Children's Traffic Fatalities in Detroit 2004-2014 (notice how the size of the dot indicates frequency). |
Topic models provide a simple way to analyze large volumes of unlabeled text. A "topic" consists of a cluster of words that frequently occur together. Using contextual clues, topic models can connect words with similar meanings and distinguish between uses of words with multiple meanings.
| Topic Modeling Tool |
| Topic Model Stacked Bar Graphs from Quantifying Kissinger. |
LXML is an awesome python tool for reading and editing xml files, and we've been using it extensively during the grant period to do programmatic cleanup to our legacy EAD files. To give an example of just how powerful the library is, late last week we ran a script to make tens of thousands of edits to all of our ~2800 EAD files, and it took all of 2 minutes to complete. This would have been an impossible task to complete manually, but lxml made it easy.
We want to share the love, so in this post we'll be walking through how we use the tool to make basic xml edits, with some exploration of the pitfalls and caveats we've encountered along the way.
Assuming you already have a version of python on your system, you'll first need to install the lxml library
In an ideal world, that should be as easy as running "pip install lxml" from a command-line. If that doesn't work, you have a few options based on what your OS is:
We'll also need an ead file (or directory of files) to work on. For this demo we'll be using the ead for the UMich gargoyle collection (raw data here).
First, we need to point lxml to the input ead.
Now we have an lxml "etree" or "element tree" object to work with, and we can do all sorts of things with it. From this parent tree, we can now select individual tags or groups of tags in the ead document to perform actions on, based on just about any criteria we can come up with. To do this we'll first need to use an "xpath" search:
There are a few things to know about lxml's xpath function:
First, it takes input in the xpath language standard, which is a standardized way to designate exact locations within an xml file. For example, the above search returns a list of every extent tag appearing in the ead file -- the double slashes at the beginning mean that it should look for those tags anywhere in the document. If I wanted to be more specific, I could use an exact search, which would be something like "/ead/archdesc/did/physdesc/extent". We will only be going into basic xpath usage here, but the language is ridiculously powerful - if you're curious as to more advanced things you can do with it, check out this tutorial.
Second, an xpath search always returns a list, even if only one result is found. It's really easy to forget this while writing a quick script, so if you're getting errors talking about your code finding a list when it didn't expect one, that's probably the reason.
A few more xpath examples:
The xpath search will give us a list of results, but to look at or edit any individual tag we'll need to grab it out of the search results. Once we have an individual element (lxml's representation of the tag) we can start to access some of its data:
Ok! Now that we know how to get at subsections of the ead file, we can start doing some programmatic edits. In our experience, our edits fall into one of just a few categories of possible changes:
We'll go through each of these and give some examples and practical tips from our own experience working with EADs at the Bentley.
This is usually a fairly straightforward task, though there is one big exception when dealing with groups of inline tags. A simple straightforward example:
This gets more complicated when you're dealing with a tag like the following:
<unittitle>Ann Arbor Township records, <unitdate>1991-2002</unitdate>, inclusive</unittitle>
Trying to access unittitle.text here will only return "Ann Arbor Township records, " and ignore everything afterwards. There is no easy way around this through lxml itself, so in these cases we've found it easiest to just convert the whole element to a string using the etree.tostring() method, doing some normal python string manipulation on that result, then converting it back into an element using etree.fromstring() and inserting it back into the ead file. That looks a little like this:
Don't worry if some of that didn't make sense -- we'll be going over more of the creating, inserting, and moving elements later on.
The most straight-forward of edits. Here's an example:
Attributes are accessed by calling .attrib on the element, which returns a python dictionary containing a set of keys (the attribute names) and their respective values:
Editing the attributes is a pretty straightforward task, largely using python's various dictionary access methods:
Here you will need to access the parent tag of the tag to be deleted using the element's .getparent() method:
There are two primary ways of going about this - one long and verbose, and the other a kind of short-hand built in to lxml. We'll do the long way first:
The alternate method is to use lxml's element builder tool. This is what that would look like:
The easiest way to do this is to treat the element objects as if they were a python list. Just like python's normal list methods, etree elements can use .insert, .append, .index, or .remove. The only gotcha to keep in mind is that lxml never copies elements when they are moved -- the singular element itself is removed from where it was and placed somewhere else. Here's a move in action:
Once you've made all the edits you want, you'll need to write the new ead data to a file. The easiest way we've found to do this is using the etree.tostring() method, but there are a few important caveats to note. .tostring() takes a few optional arguments you will want to be sure to include: to keep your original xml declaration you'll need to set xml_declaration=True, and to keep a text encoding statement, you'll need encoding="utf-8" (or whatever encoding you're working with):
We can also pretty-print the results, which will help ensure the ead file has well-formed indentation, and is generally not an incomprehensible mess of tags. Because of some oddities in the way lxml handles tag spacing, to get pretty-print to work you'll need to add one extra step to the the input file parsing process:
Note that the new parser will effectively remove all whitespace (spaces and newlines) between tags, which can cause problems if you have any complicated tag structure. We had some major issues with this early on, and ended up writing our own custom pretty-printing code on top of what is already in lxml, which ensures that inline tags keep proper spacing (as in, <date>1926,</date> <date>1965</date> doesn't become <date>1926,</date><date>1965</date>), and to prevent other special cases like xml lists from collapsing into big blocks of tags. Anyone is welcome to use or adapt what we've written - check it out here!
Thanks for reading along! We've found lxml to be indispensable in our cleanup work here at the Bentley, and we hope you'll find it useful as well. And if you have any thoughts or use any other tools in your own workflows we'd love to hear about them -- let us know in the comments below!
In this context, the term “provenance” connotes the individual, family, or organization that created or received the items in a collection. The principle of provenance or the respect des fonds dictates that records of different origins (provenance) be kept separate to preserve their context.
In this context, the term “original order” connotes the organization and sequence of records established by the creator of the records.
"Digital materials are especially vulnerable to loss and destruction because they are stored on fragile magnetic and optical media that deteriorate rapidly and that can fail suddenly." (Hedstrom and Montgomery 1998)
"Unlike the situation that applies to books, digital archiving requires relatively frequent investments to overcome rapid obsolescence introduced by galloping technological change." (Feeney 1999)
“While it is technically feasible to alter records in a paper environment, the relative ease with which this can be achieved in the digital environment, either deliberately or inadvertently, has given this issue more pressing urgency.”
@meau nope...that's why i don't see the logic behind virtual reading rooms...we're creating unnecessary impediments to access.
— Jarrett M. Drake (@jmddrake) August 10, 2015